According to data from the World Health Organization (WHO), breast cancer is the second most commonly diagnosed cancer globally, after lung cancer, and remains the leading cause of cancer incidence and mortality among women.
The evolution of HER2-targeted therapy has progressed through multiple stages. In 2019, the antibody-drug conjugate (ADC) T-DXd (Trastuzumab Deruxtecan, Enhertu) was approved, offering a new strategy to overcome resistance to the previous generation ADC drug, T-DM1. The cytotoxic payload, DXd, carried by this drug is membrane-permeable. This property allows it not only to precisely target HER2-high-expressing cancer cells but also to eliminate adjacent HER2-negative tumor cells through the "bystander effect," resulting in superior efficacy. However, as clinical use expands, acquired resistance inevitably emerges in some patients. Consequently, establishing reliable preclinical resistance models has become crucial for advancing subsequent research.
Current strategies for constructing resistance models primarily fall into four categories (Figure 1):
Patient-Derived Xenograft (PDX) Models: These models transplant resistant tumor tissue from patients into immunodeficient mice. They effectively preserve tumor heterogeneity and are suitable for individualized therapy research, but face challenges such as long modeling cycles and limited reproducibility.
In Vivo Induced Resistance: This method simulates the evolution of resistance by inoculating sensitive tumor cells into mice and repeatedly administering the drug. It closely mirrors the real physiological environment, but the overall cycle remains relatively long.
In Vitro Induced Resistance: This approach involves gradually increasing drug concentration in the cell culture medium to select for resistant cell lines under drug pressure. It is convenient and has a short cycle, but carries the risk of insufficient correlation between in vitro and in vivo results.
Gene-Edited Resistance Models: This technique utilizes gene-editing technology to introduce specific resistance mutations, allowing for the precise study of single mechanisms. However, it struggles to replicate the multi-gene, multi-pathway complexity of clinical resistance.

Figure 1. Methods for Constructing Resistance Models in Preclinical Research
To address the lack of models for Enhertu resistance research, GemPharmatech employed a combined in vivo and in vitro induction strategy, successfully establishing a stable JIMT-1-Enhertu-R resistance model in BALB/c-Nude mice.
Further in vivo animal experiments confirmed that tumors in this model showed no significant inhibition under treatment with 3 mg/kg Enhertu (Figure 2).

Figure 2. In Vivo Resistance Validation of JIMT-1-Enhertu-R
According to literature reports, Enhertu resistance mechanisms are multi-level and involve intertwined factors. These include ADC-payload resistance mutations, alterations in HER2 target expression, as well as processes involving drug delivery, intracellular drug processing, activation of drug efflux pumps, activation of bypass signaling pathways, and immune escape within the tumor microenvironment. Therefore, after obtaining the Enhertu-resistant phenotype, GemPharmatech conducted preliminary investigations into its potential resistance mechanisms:
In Vitro Cell Killing Assays demonstrated that JIMT-1-Enhertu-R cells exhibited significantly increased tolerance to the free toxin DXd, while retaining sensitivity to MMAE (Figure 3).
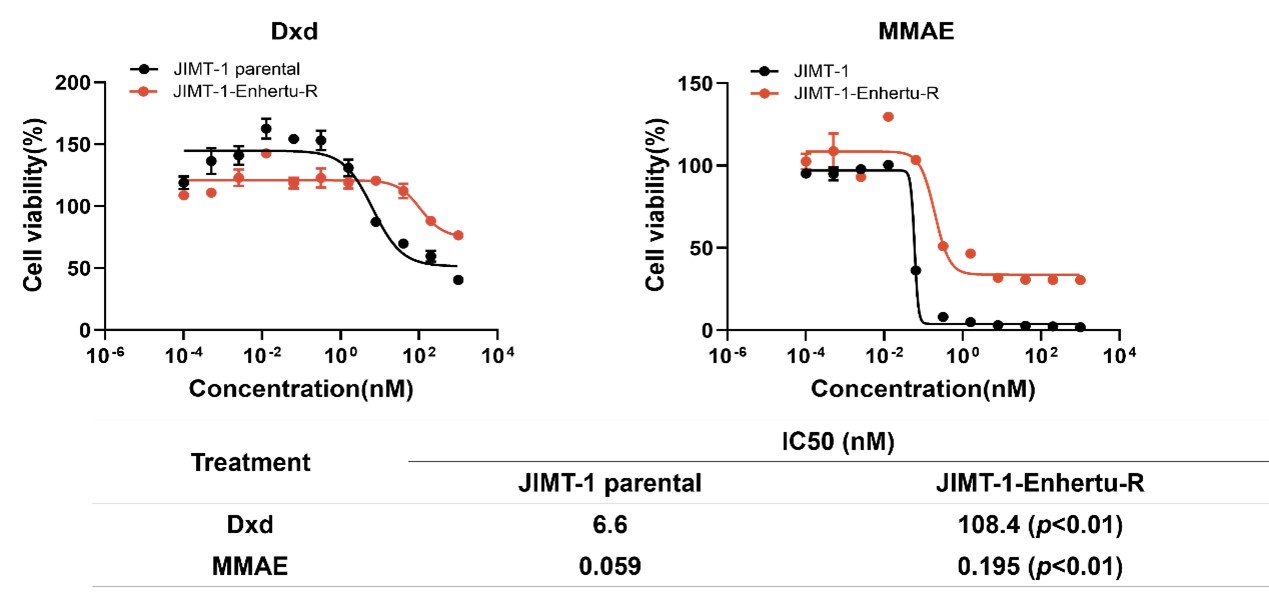
Figure 3. IC50 Determination of Different Toxins in JIMT-1 Parental and JIMT-1-Enhertu-R Cells
Flow Cytometry analysis of HER2 expression showed no significant decrease in HER2 levels in either the JIMT-1-Enhertu-R cell line or the corresponding tumor masses (Figure 4).

Figure 4. HER2 Expression Profiling in In Vitro and In Vivo JIMT-1 and JIMT-1-Enhertu-R Models
Immunohistochemistry (IHC) results of tissue sections indicated that, compared to the JIMT-1 parental line, HER2 expression in the resistant model tumor tissue was uniform and stable (Figure 5).

Figure 5. IHC Staining Results for HER2 in JIMT-1 WT and JIMT-1-Enhertu-R Tumor Tissues
Based on these collective results, we hypothesize that the resistance mechanism in this model may be related to reduced cellular sensitivity to the payload, rather than being caused by target loss. This finding also suggests a theoretical basis for potential cross-resistance to other ADC drugs sharing similar payloads.
GemPharmatech has also established an "Enhanced" version of the Enhertu resistance model, which maintains good tolerance to the drug even at high doses (Figure 6). This provides robust model support for research aiming to overcome Enhertu efficacy and for new drug testing.

Figure 6. Efficacy Test of High-Dose Enhertu in the JIMT-1-Enhertu-R Enhanced Model
As Enhertu sees widespread clinical use and gradually expands into earlier lines of therapy, its potential resistance issues are becoming increasingly prominent. As a result, our resistance models are useful not only for screening novel drugs, optimizing ADC structures, and developing combination therapies, but also are indispensable tools for deciphering the underlying biological mechanisms of resistance.
At GemPharmatech, we have established various tumor resistance model platforms covering multiple areas, including resistance to immune checkpoint inhibitors (e.g., PD-1), ADC drug-induced resistance, and EGFR-TKI resistance. These modeling approaches encompass spontaneous resistance, in vivo and in vitro induction, and gene editing, among other technical pathways. Some models are already available for external applications, while others are under continuous development to support the advancement of innovative anti-tumor drug research.
Anti-PD1 Resistance Models:
Induced Acquired Resistant Models | Gene Editing Acquired Resistant Models | Primary Resistant Models | |
Cell lines | Keytruda induced CT26-PD1-R, MC38-PD1-R | CT26-B2M KO | KPC, Pan02, B16F10, LLC, 4T1, EMT6 , RENCA, MB49, MOPC315, MPC-11 |
Features | Match clinical symptoms The drug resistance mechanism is complex | Close to clinical symptoms The drug resistance mechanism is clear | Match clinical symptoms The drug resistance mechanism is complex |
Application | • New PD1 monoclonal antibody drug testing • Chemotherapy drug testing • Activate other immune cells | • Mechanistically defined drug development | • New PD1 monoclonal antibody drug testing • Chemotherapy drug testing • Activate other immune cells |
GemPharmatech's Resistant Models:

Contact us at sales@gempharmatech.com to learn more!